Prior Distributions with Bilby
Prior distributions are a core component of any Bayesian problem and specifying them in codes can be one of the most confusing elements of a code. The prior modules in Bilby provide functionality for specifying prior distributions in a natural way.
We have a range of predefined types of prior distribution and each kind has methods to:
draw samples,
prior.sample.calculate the prior probability,
prior.prob.rescale samples from a unit cube to the prior distribution,
prior.rescale. This is especially useful when using nested samplers as it avoids the need for rejection sampling.Calculate the log prior probability,
prior.log_prob.
In addition to the predefined prior distributions there is functionality to specify your own prior, either from a pair of arrays, or from a file.
Each prior distribution can also be given a name and may have a different latex_label for plotting. If no name is provided, the default is None (this should probably by '').
[1]:
import bilby
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
Prior Instantiation
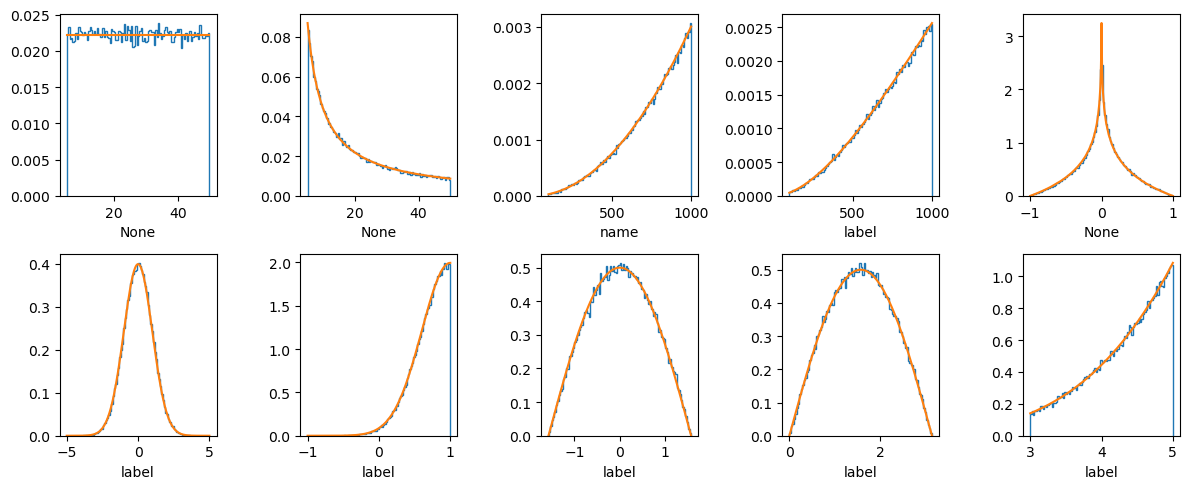
Below we demonstrate instantiating a range of prior distributions.
Note that when a latex_label is not specified, the name is used.
[2]:
fig = plt.figure(figsize=(12, 5))
priors = [
bilby.core.prior.Uniform(minimum=5, maximum=50),
bilby.core.prior.LogUniform(minimum=5, maximum=50),
bilby.core.prior.PowerLaw(name="name", alpha=2, minimum=100, maximum=1000),
bilby.gw.prior.UniformComovingVolume(
name="luminosity_distance", minimum=100, maximum=1000, latex_label="label"
),
bilby.gw.prior.AlignedSpin(),
bilby.core.prior.Gaussian(name="name", mu=0, sigma=1, latex_label="label"),
bilby.core.prior.TruncatedGaussian(
name="name", mu=1, sigma=0.4, minimum=-1, maximum=1, latex_label="label"
),
bilby.core.prior.Cosine(name="name", latex_label="label"),
bilby.core.prior.Sine(name="name", latex_label="label"),
bilby.core.prior.Interped(
name="name",
xx=np.linspace(0, 10, 1000),
yy=np.linspace(0, 10, 1000) ** 4,
minimum=3,
maximum=5,
latex_label="label",
),
]
for ii, prior in enumerate(priors):
fig.add_subplot(2, 5, 1 + ii)
plt.hist(prior.sample(100000), bins=100, histtype="step", density=True)
if not isinstance(prior, bilby.core.prior.Gaussian):
plt.plot(
np.linspace(prior.minimum, prior.maximum, 1000),
prior.prob(np.linspace(prior.minimum, prior.maximum, 1000)),
)
else:
plt.plot(np.linspace(-5, 5, 1000), prior.prob(np.linspace(-5, 5, 1000)))
plt.xlabel("{}".format(prior.latex_label))
plt.tight_layout()
plt.show()
plt.close()
Define an Analytic Prior
Adding a new analytic is achieved as follows.
[3]:
class Exponential(bilby.core.prior.Prior):
"""Define a new prior class where p(x) ~ exp(alpha * x)"""
def __init__(self, alpha, minimum, maximum, name=None, latex_label=None):
super(Exponential, self).__init__(
name=name, latex_label=latex_label, minimum=minimum, maximum=maximum
)
self.alpha = alpha
def rescale(self, val):
return (
np.log(
(np.exp(self.alpha * self.maximum) - np.exp(self.alpha * self.minimum))
* val
+ np.exp(self.alpha * self.minimum)
)
/ self.alpha
)
def prob(self, val):
in_prior = (val >= self.minimum) & (val <= self.maximum)
return (
self.alpha
* np.exp(self.alpha * val)
/ (np.exp(self.alpha * self.maximum) - np.exp(self.alpha * self.minimum))
* in_prior
)
[4]:
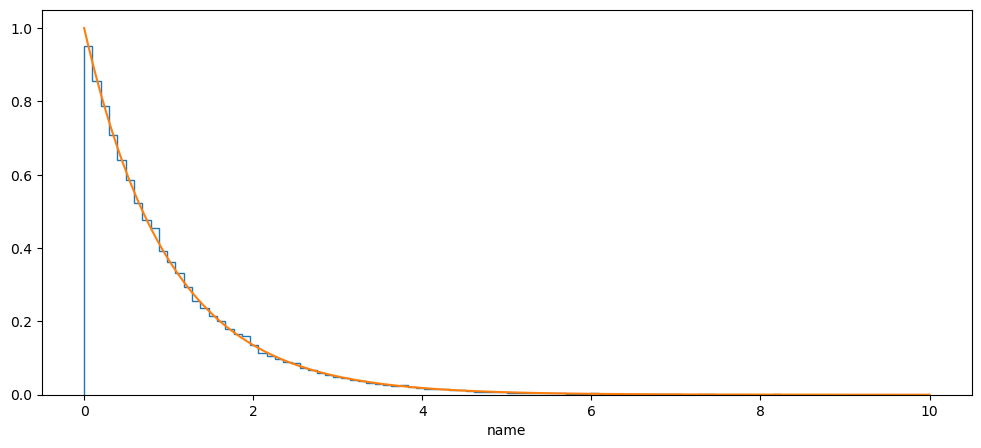
prior = Exponential(name="name", alpha=-1, minimum=0, maximum=10)
plt.figure(figsize=(12, 5))
plt.hist(prior.sample(100000), bins=100, histtype="step", density=True)
plt.plot(
np.linspace(prior.minimum, prior.maximum, 1000),
prior.prob(np.linspace(prior.minimum, prior.maximum, 1000)),
)
plt.xlabel("{}".format(prior.latex_label))
plt.show()
plt.close()